Research Interests
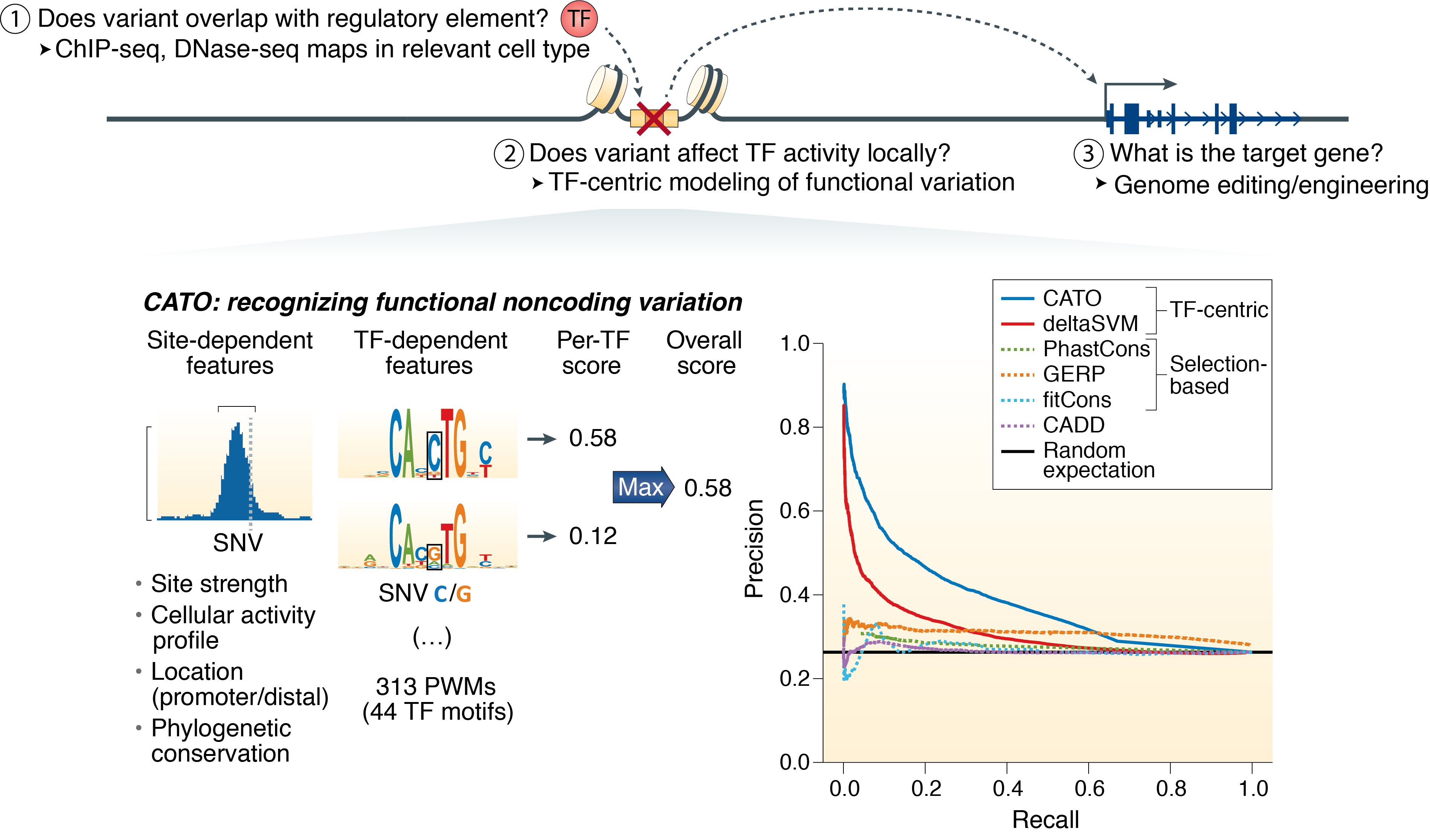
The torrent of variants emerging from human resequencing studies coupled with the growing number of common, disease-associated noncoding variants has created an urgent need for determining the consequences of variation within regulatory DNA. In contrast to the vast diversity of protein function, the elements that regulate gene expression recruit from a shared repertoire of transcription factors, offering the potential for a common regulatory sequence code. Our lab is thus interested in establishing a foundation for the mechanistic study of regulatory variation, with the long-term goal of understanding how the noncoding genome affects cell-type specific transcriptional regulation. We apply both experimental and computational approaches and our interests include:
- Genome engineering approaches for dissection of regulatory architecture
- Prediction of functional effects of non-coding regulatory variation
- Regulatory variation and the genetics of human diseases and traits
- Profiling of chromatin features and their relationship to nuclear organization and function